STRUMPACK vs EIGEN performance
The goal of this notebook is to allow the documentation of STRUMPACK vs EIGEN performance to be maintained in a single accessible location. The environment within which this notebook is run follows the standard cctbx conda build instructions available here. For this instance, we are using the STRUMPACK-enabled build of cctbx located at ExaFEL:cctbx_project(str_merge). STRUMPACK is currently built using the installation script STRUMPACK_installer_shared.sh, and if the installation takes place within the same directory as moddules and build, the cctbx build process can make use of it as a backend. After the STRUMPACK solver finishes, enter the build directory, run libtbx.refresh and make. The STRUMPACK-supported modules should now build and link with the new backend.
The solution of the below systems assume that we have some A and b data already, and we can provide these in CSV file format to the solver (A is in “row col value” format). We can begin by importing all of the required modules. We make use of Numpy’s ability to parse CSV files into numeric format, and SciPy’s sparse storage format to format the matrices into the required CSR sparse storage format for solving.
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
import matplotlib
import matplotlib.pyplot as plt
font = {'family' : 'serif',
#'weight' : 'bold',
'size' : 12}
matplotlib.rc('font', **font)
matplotlib.rcParams['figure.figsize'] = (16,10)
matplotlib.rcParams['figure.dpi']= 150
import matplotlib.gridspec as gridspec
from __future__ import division
from cctbx.array_family import flex
from libtbx.test_utils import approx_equal
from libtbx.development.timers import Profiler
import sys
import numpy as np
import scipy.sparse as sps
%env BOOST_ADAPTBX_FPE_DEFAULT=1
%env BOOST_ADAPTBX_SIGNALS_DEFAULT=1
env: BOOST_ADAPTBX_FPE_DEFAULT=1
env: BOOST_ADAPTBX_SIGNALS_DEFAULT=1
We load the A and b data set for solving from the locations specified below. Ideally, we can loop over and load all different data sets and process each individually. For a working example, we begin by processing a single data set, comparing Eigen and Strumpack for varying levels of shared parallelism.
A_path="/net/dials/raid1/mlxd/InDev/feb_sprint/sam/samosa/step4K_samosa_debug_1k/out_strumpack_1k_omp1_paramslevmar.parameter_flags=Bfactor-/A_strum_1k_omp1_paramslevmar.parameter_flags=Bfactor-.csv"
A_mat = np.loadtxt(A_path,dtype={'names':('rows','cols','vals'),'formats':('i8','i8','f8')})
b_path="/net/dials/raid1/mlxd/InDev/feb_sprint/sam/samosa/step4K_samosa_debug_1k/out_strumpack_1k_omp1_paramslevmar.parameter_flags=Bfactor-/b_strum_1k_omp1_paramslevmar.parameter_flags=Bfactor-.csv"
b_vec = np.loadtxt(b_path)
We are solving system of normal equations, and so we can safely assume that we have a square matrix. This ensures the number of rows and colums will be identical, which can be read from the length of the numpy array b_vec.
n_rows = len(b_vec)
n_cols = n_rows
We next wish to create a SciPy CSR sparse matrix type using the data from A_mat.
A_sp = sps.csr_matrix((A_mat['vals'],(A_mat['rows'],A_mat['cols'])))
With this, we can now create the appropriate flex data types for solving the linear system. While the operations performed upto and including this point involve many copy steps, it can be stated that the time to solve the system is much greater than the time to perform internal copies, and so we can ignore the times required for these steps for any realistic datasets.
For the solvers we require the row index pointers, the column indices, and sparse matrix values. These can be read directly and converted to flex types as follows.
A_indptr = flex.int(A_sp.indptr)
A_indices = flex.int(A_sp.indices)
A_values = flex.double(A_sp.data)
b = flex.double(b_vec)
With the data in the required format, we can now create a solver object and solve the system. We also include a built-in profiler object to time this solver step for both the STRUMPACK and EIGEN backends.
To keep a record of the timings, we can create a dictionary to store each respective value.
import time
timing_dict = {"strum":0, "eigen":0}
Next, we load the STRUMPACK-enabled solver module, and define the function to call the solver. This solver code is contained within “modules/cctbx_project/scitbx/examples/bevington/strumpack_solver_ext.cpp” and creates a Boost.Python wrapper for both an EIGEN and STRUMPACK Ax=b solver object.
A non-notebook script exists to test this functionality at “modules/cctbx_project/scitbx/examples/bevington/strumpack_eigen_solver.py”, which can be called as:
libtbx.python strumpack_eigen_solver.py A_mat.csv b_vec.csv
where A_mat.csv and b_vec.csv are the CSV files with the linear system to be solved.
In this notebook, we can instead load and process the files explicitly.
import scitbx_examples_strumpack_solver_ext as solver_ext
es = solver_ext.eigen_solver
ss = solver_ext.strumpack_solver
def run_solver(n_rows, n_cols, A_indptr ,A_indices, A_values, b):
P = Profiler("STRUMPACK")
#timing_dict["strum"]=time.time()
res_strum = ss(n_rows, n_cols, A_indptr, A_indices, A_values, b)
#timing_dict["strum"]=time.time()-timing_dict["strum"]
del P
P = Profiler("EIGEN")
#timing_dict["eigen"]=time.time()
res_eig = es(n_rows, n_cols, A_indptr, A_indices, A_values, b)
#timing_dict["eigen"]=time.time() - timing_dict["eigen"]
del P
for i in xrange(len(res_strum.x)):
assert( approx_equal(res_strum.x[i], res_eig.x[i]) )
run_solver(n_rows, n_cols, A_indptr ,A_indices, A_values, b)
individual call time for STRUMPACK: CPU, 291.570s; elapsed, 5.425s
individual call time for EIGEN: CPU, 5.950s; elapsed, 0.266s
While we can use the above method to process the data, we must respecify the OMP_NUM_THREADS variable to observe different levels of scalability. %env OMP_NUM_THREADS is sufficient to test this for the first run, however for an undetermined reason the notebook must be started to allow the value to be changed. There, we can run the above tasks from a !<command> cell, specifying the thread count here instead.
OMP_SOLVER='''
from __future__ import division
from cctbx.array_family import flex
from libtbx.test_utils import approx_equal
from libtbx.development.timers import Profiler
import sys
import numpy as np
import scipy.sparse as sps
import matplotlib
import matplotlib.pyplot as plt
import matplotlib
import matplotlib.pyplot as plt
matplotlib.rcParams['figure.figsize'] = (16,10)
A_path=sys.argv[1]
A_mat = np.loadtxt(A_path,dtype={'names':('rows','cols','vals'),'formats':('i8','i8','f8')})
b_path=sys.argv[2]
b_vec = np.loadtxt(b_path)
n_rows = len(b_vec)
n_cols = n_rows
A_sp = sps.csr_matrix((A_mat['vals'],(A_mat['rows'],A_mat['cols'])))
A_indptr = flex.int(A_sp.indptr)
A_indices = flex.int(A_sp.indices)
A_values = flex.double(A_sp.data)
b = flex.double(b_vec)
#import time
#timing_dict = {"strum":0, "eigen":0}
import scitbx_examples_strumpack_solver_ext as solver_ext
es = solver_ext.eigen_solver
ss = solver_ext.strumpack_solver
def run_solver(n_rows, n_cols, A_indptr ,A_indices, A_values, b):
P = Profiler("STRUMPACK")
#timing_dict["strum"]=time.time()
res_strum = ss(n_rows, n_cols, A_indptr, A_indices, A_values, b)
#timing_dict["strum"]=time.time()-timing_dict["strum"]
del P
P = Profiler("EIGEN")
#timing_dict["eigen"]=time.time()
res_eig = es(n_rows, n_cols, A_indptr, A_indices, A_values, b)
#timing_dict["eigen"]=time.time() - timing_dict["eigen"]
del P
for i in xrange(len(res_strum.x)):
assert( approx_equal(res_strum.x[i], res_eig.x[i]) )
run_solver(n_rows, n_cols, A_indptr ,A_indices, A_values, b)
'''
OMP_SOLVER = OMP_SOLVER
OMP_SOLVER_FILE = open("OMP_SOLVER.py", "w")
OMP_SOLVER_FILE.write(OMP_SOLVER)
OMP_SOLVER_FILE.close()
DATAPATH="/net/dials/raid1/mlxd/InDev/feb_sprint/sam/samosa/"
A_LIST = !find {DATAPATH} -iname "A*.csv"
B_LIST = [ii.replace('/A_','/b_') for ii in A_LIST]
We now record the indices of the data with a specific number of images. We can use these indices to later submit jobs with a given number of frames, and hence resulting matrix size.
list_idx={}
for imgs in ['1k','5k','10k','32k']:
list_idx.update({imgs:[i for i, j in enumerate(A_LIST) if imgs in j]})
list_idx
{'10k': [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14],
'1k': [15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29],
'32k': [30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44],
'5k': [45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59]}
With the file writtent to disk, we now create a loop in mixed Python/Bash to call the solver, then extract the standard output from the profiler to examine the scalability. The profiler code wraps each solver only, and does not take into account the load-times for the data.
str_out={}
import os
threads_list = [1,2,4,8,16,32,64]
for imgs_size in list_idx:
print "Data Set Size:=%s"%imgs_size
#Subselect the smallest data size for now
if imgs_size != "1k":
print "Skipping %s"%imgs_size
continue
for imgs_idx in list_idx[imgs_size]:
dat_name = A_mat.split('/')[-1][2:-4]
print "Data Set Name:=%s"%dat_name
for threads in threads_list:
print "OMP_NUM_THREADS:=%d"%threads
A_mat = A_LIST[imgs_idx]; b_vec = B_LIST[imgs_idx]
#Ensure the A and b data are matched correctly
assert(os.path.dirname(A_mat) == os.path.dirname(b_vec))
val = !OMP_NUM_THREADS={threads} libtbx.python ./OMP_SOLVER.py {A_mat} {b_vec}
key = 'omp' + str(threads) + '_' + dat_name
str_out.update({key:val})
—CLICK FOR OUTPUT—
Data Set Size:=10k
Skipping 10k
Data Set Size:=1k
Data Set Name:=strum_1k_omp1_paramslevmar.parameter_flags=Rxy-levmar.parameter_flags=Bfactor-levmar.parameter_flags=Deff-levmar.parameter_flags=Eta-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_1k_omp1_paramslevmar.parameter_flags=Rxy-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_1k_omp1_paramslevmar.parameter_flags=Bfactor-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_1k_omp1_paramslevmar.parameter_flags=Deff-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_1k_omp1_paramslevmar.parameter_flags=Eta-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_1k_omp1_paramslevmar.parameter_flags=Rxy-levmar.parameter_flags=Bfactor-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_1k_omp1_paramslevmar.parameter_flags=Rxy-levmar.parameter_flags=Deff-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_1k_omp1_paramslevmar.parameter_flags=Rxy-levmar.parameter_flags=Eta-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_1k_omp1_paramslevmar.parameter_flags=Bfactor-levmar.parameter_flags=Deff-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_1k_omp1_paramslevmar.parameter_flags=Bfactor-levmar.parameter_flags=Eta-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_1k_omp1_paramslevmar.parameter_flags=Deff-levmar.parameter_flags=Eta-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_1k_omp1_paramslevmar.parameter_flags=Rxy-levmar.parameter_flags=Bfactor-levmar.parameter_flags=Deff-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_1k_omp1_paramslevmar.parameter_flags=Rxy-levmar.parameter_flags=Bfactor-levmar.parameter_flags=Eta-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_1k_omp1_paramslevmar.parameter_flags=Rxy-levmar.parameter_flags=Deff-levmar.parameter_flags=Eta-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_1k_omp1_paramslevmar.parameter_flags=Bfactor-levmar.parameter_flags=Deff-levmar.parameter_flags=Eta-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Size:=5k
Skipping 5k
Data Set Size:=32k
Skipping 32k
In the event of a crash, it is wise to save the resulting data (JSON and Pickle) to disk…
import json, cPickle
with open("OMP_SOLVER_TIME_1k.json", "w") as OMP_SOLVER_TIME_FILE:
OMP_SOLVER_TIME_FILE.write(json.dumps(str_out))
with open("OMP_SOLVER_TIME_1k.pickle", "w") as OMP_SOLVER_TIME_FILE:
OMP_SOLVER_TIME_FILE.write(cPickle.dumps(str_out))
…which can then be easily reloaded later. Now, we parse the string, extract the timing data, and add it to the python object at the specified thread number index. This allows for easily sorting and plot the data later.
import cPickle
str_out=cPickle.load(open('OMP_SOLVER_TIME_1k.pickle', 'rb'))
To simplify indexing later for plots, we create shortened keys. Additionally, we ca also separate the data based upon unique index set and thread numbers.
str_out_sm={}
for k in str_out.keys():
str_out_sm.update({k.replace('levmar.parameter_flags=','').replace('_strum_1k_omp1_params','_'):str_out[k]})
u_dat={}
threads_list=[1,2,4,8,16,32,64]
uniq_ref = set([k.split('_')[1] for k in str_out_sm.keys()])
df_list=[]
for u in uniq_ref:
same_t = lambda: None #Functions are objects, so legit
same_t.strum = []
same_t.eig = []
for t in threads_list:
same_t.strum.append(str_out_sm["omp%d_%s"%(t,u)][0].rsplit(',')[-1].strip()[0:-1])
same_t.eig.append(str_out_sm["omp%d_%s"%(t,u)][1].rsplit(',')[-1].strip()[0:-1])
u_dat.update({u:same_t})
str_E = "EIG_%s"%u
str_S = "STRUM_%s"%u
df_list.append( pd.DataFrame({str_E:same_t.eig,str_S:same_t.strum}, index=threads_list).transpose() )
We now have a list of Pandas Dataframes, with which we can combine to a single entity. We can then proceed to plot the resulting columns, comparing both Eigen and STRUMPACK for each specific set of refined parameters.
import pandas as pd
perf_s = pd.concat(df_list).transpose()
perf_s
—CLICK FOR OUTPUT—
| EIG_Rxy-Deff-Eta- | STRUM_Rxy-Deff-Eta- | EIG_Bfactor- | STRUM_Bfactor- | EIG_Bfactor-Deff-Eta- | STRUM_Bfactor-Deff-Eta- | EIG_Rxy-Bfactor-Deff-Eta- | STRUM_Rxy-Bfactor-Deff-Eta- | EIG_Deff-Eta- | STRUM_Deff-Eta- | ... | EIG_Eta- | STRUM_Eta- | EIG_Deff- | STRUM_Deff- | EIG_Rxy-Bfactor- | STRUM_Rxy-Bfactor- | EIG_Rxy-Bfactor-Eta- | STRUM_Rxy-Bfactor-Eta- | EIG_Rxy-Bfactor-Deff- | STRUM_Rxy-Bfactor-Deff- | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.013 | 15.598 | 0.010 | 12.359 | 0.020 | 20.328 | 0.011 | 20.885 | 0.018 | 21.803 | ... | 0.017 | 22.696 | 0.051 | 10.401 | 0.017 | 20.001 | 0.016 | 18.391 | 0.018 | 18.415 |
| 2 | 0.013 | 8.859 | 0.011 | 7.880 | 0.021 | 11.665 | 0.013 | 11.145 | 0.019 | 11.933 | ... | 0.016 | 10.479 | 0.053 | 5.888 | 0.018 | 12.719 | 0.019 | 10.128 | 0.019 | 9.713 |
| 4 | 0.012 | 5.314 | 0.009 | 4.655 | 0.020 | 8.141 | 0.013 | 7.335 | 0.019 | 7.673 | ... | 0.018 | 6.631 | 0.057 | 3.791 | 0.017 | 7.902 | 0.019 | 6.261 | 0.019 | 5.929 |
| 8 | 0.012 | 3.838 | 0.015 | 3.328 | 0.020 | 4.839 | 0.016 | 3.587 | 0.028 | 5.963 | ... | 0.016 | 5.231 | 0.056 | 2.671 | 0.015 | 4.368 | 0.015 | 4.427 | 0.016 | 4.156 |
| 16 | 0.018 | 3.186 | 0.015 | 2.485 | 0.022 | 4.720 | 0.020 | 3.667 | 0.032 | 4.466 | ... | 0.016 | 6.855 | 0.059 | 2.156 | 0.018 | 4.799 | 0.021 | 3.579 | 0.017 | 3.526 |
| 32 | 0.020 | 3.269 | 0.011 | 2.776 | 0.041 | 4.463 | 0.025 | 3.016 | 0.021 | 3.709 | ... | 0.026 | 3.214 | 0.087 | 2.134 | 0.034 | 4.253 | 0.021 | 4.905 | 0.019 | 3.712 |
| 64 | 0.016 | 6.079 | 0.026 | 4.142 | 0.027 | 4.427 | 0.015 | 5.049 | 0.027 | 5.785 | ... | 0.023 | 4.746 | 0.086 | 3.149 | 0.023 | 6.350 | 0.021 | 6.270 | 0.032 | 5.650 |
7 rows × 30 columns
With the performance data in an easily accessible format, we can plot each respective data set for both solvers. Each title corresponds to the parameters being refined, the x-axis is the number of OpenMP threads used in the solution, and the y-axis is the time to solve the system in seconds.
# define the figure size and grid layout properties
figsize = (10, 8)
cols = 4
gs = gridspec.GridSpec(cols, cols)
gs.update(hspace=0.6)
fig1 = plt.figure(num=1, figsize=figsize)
fig1.suptitle('1k data', size=16)
ax = []
for i, u in enumerate(uniq_ref):
row = (i // cols)
col = i % cols
ax.append(fig1.add_subplot(gs[row, col]))
if(col==0):
ax[-1].set_ylabel('time [s]')
ax[-1].set_title(str(u)[:-1])
ax[-1].plot(perf_s.index, perf_s['EIG_'+u], 'o', ls='-', ms=6, label='EIG')
ax[-1].plot(perf_s.index, perf_s['STRUM_'+u], 'x', ls='-', ms=6, label='STRUM')
ax[-1].xaxis.set_ticks(threads_list)
ax[-1].set_xscale('log', basex=2)
ax[-1].set_xlim([threads_list[0]/1.8,threads_list[-1]*1.8])
ymin, ymax = ax[-1].get_ylim()
ax[-1].set_ylim([-1,ymax+1])
for label in ax[-1].yaxis.get_ticklabels()[::2]:
label.set_visible(False)
for label in ax[-1].xaxis.get_ticklabels()[1::2]:
label.set_visible(False)
ax[-1].legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.,)
<matplotlib.legend.Legend at 0x7fc4eada0d50>
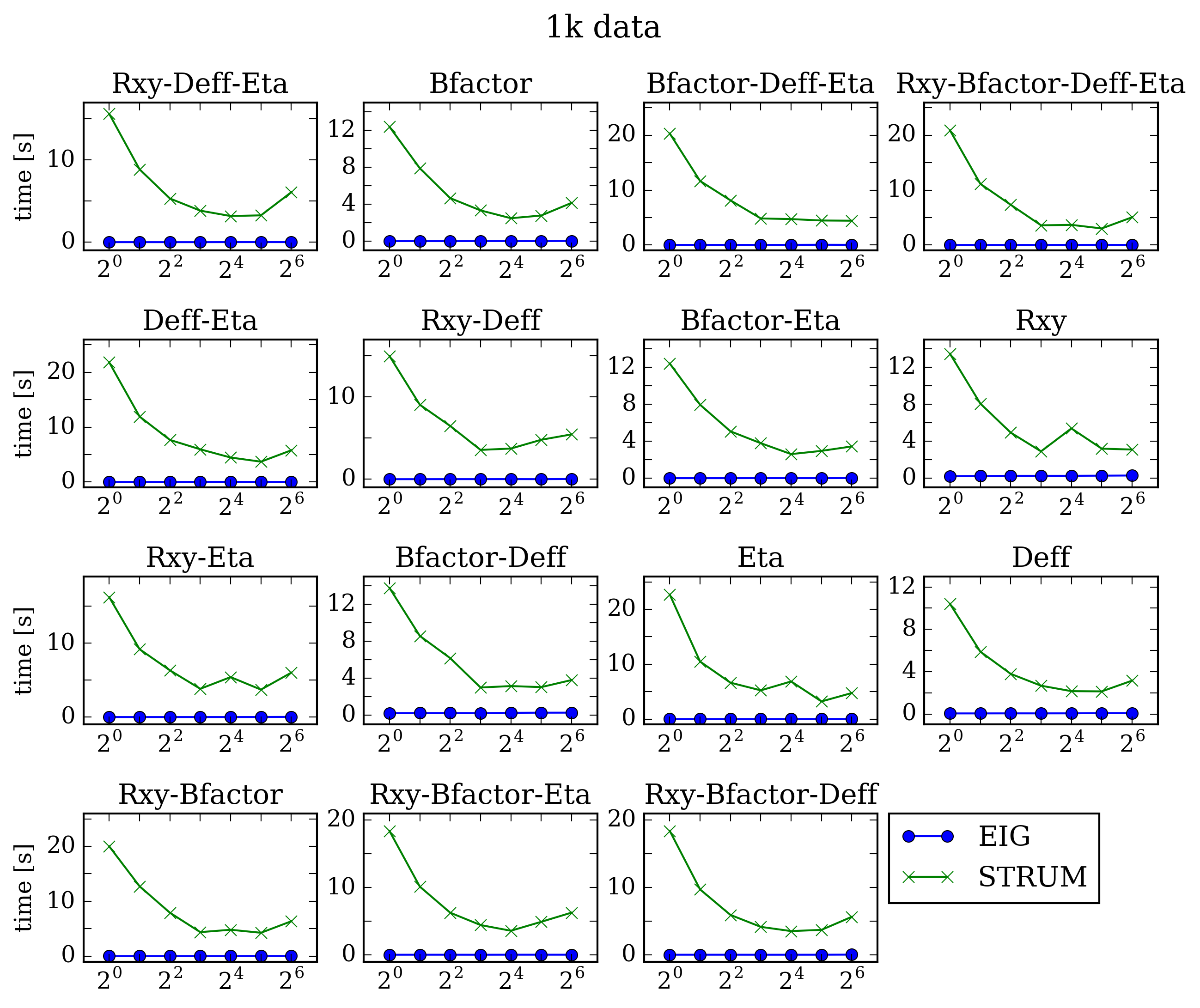 |
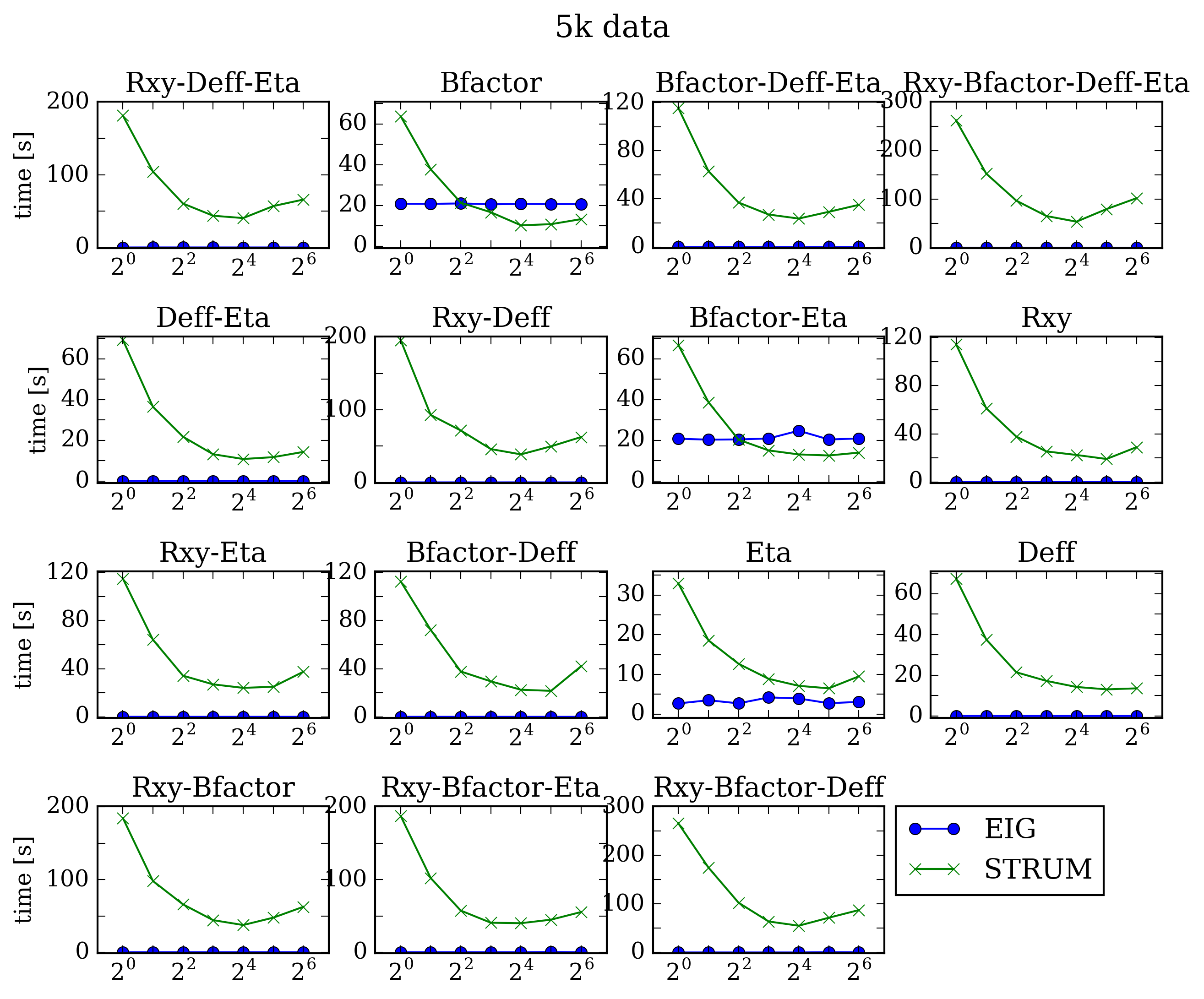
As expected, the OMP_NUM_THREADS variable allowed the STRUMPACK code to improve in performance, but for this data size EIGEN clearly offers better performance. Next we increase the number of frames in the dataset by a factor of 5, and perform the same analysis.
import os
threads_list = [1,2,4,8,16,32,64]
for imgs_size in list_idx:
print "Data Set Size:=%s"%imgs_size
#Subselect the smallest data size for now
if imgs_size != "5k":
print "Skipping %s"%imgs_size
continue
for imgs_idx in list_idx[imgs_size]:
A_mat = A_LIST[imgs_idx]; b_vec = B_LIST[imgs_idx]
dat_name = A_mat.split('/')[-1][2:-4]
print "Data Set Name:=%s"%dat_name
#Ensure the A and b data are matched correctly
assert(os.path.dirname(A_mat) == os.path.dirname(b_vec))
for threads in threads_list:
print "OMP_NUM_THREADS:=%d"%threads
val = !OMP_NUM_THREADS={threads} libtbx.python ./OMP_SOLVER.py {A_mat} {b_vec}
key = 'omp' + str(threads) + '_' + dat_name
str_out.update({key:val})
import json, cPickle
with open("OMP_SOLVER_TIME_5k.json", "w") as OMP_SOLVER_TIME_FILE:
OMP_SOLVER_TIME_FILE.write(json.dumps(str_out))
with open("OMP_SOLVER_TIME_5k.pickle", "w") as OMP_SOLVER_TIME_FILE:
OMP_SOLVER_TIME_FILE.write(cPickle.dumps(str_out))
—CLICK FOR OUTPUT—
Data Set Size:=10k
Skipping 10k
Data Set Size:=1k
Skipping 1k
Data Set Size:=5k
Data Set Name:=strum_5k_omp1_paramslevmar.parameter_flags=Rxy-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_5k_omp1_paramslevmar.parameter_flags=Bfactor-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_5k_omp1_paramslevmar.parameter_flags=Deff-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_5k_omp1_paramslevmar.parameter_flags=Eta-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_5k_omp1_paramslevmar.parameter_flags=Rxy-levmar.parameter_flags=Bfactor-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_5k_omp1_paramslevmar.parameter_flags=Rxy-levmar.parameter_flags=Deff-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_5k_omp1_paramslevmar.parameter_flags=Rxy-levmar.parameter_flags=Eta-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_5k_omp1_paramslevmar.parameter_flags=Bfactor-levmar.parameter_flags=Deff-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_5k_omp1_paramslevmar.parameter_flags=Bfactor-levmar.parameter_flags=Eta-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_5k_omp1_paramslevmar.parameter_flags=Deff-levmar.parameter_flags=Eta-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_5k_omp1_paramslevmar.parameter_flags=Rxy-levmar.parameter_flags=Bfactor-levmar.parameter_flags=Deff-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_5k_omp1_paramslevmar.parameter_flags=Rxy-levmar.parameter_flags=Bfactor-levmar.parameter_flags=Eta-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_5k_omp1_paramslevmar.parameter_flags=Rxy-levmar.parameter_flags=Deff-levmar.parameter_flags=Eta-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_5k_omp1_paramslevmar.parameter_flags=Bfactor-levmar.parameter_flags=Deff-levmar.parameter_flags=Eta-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Name:=strum_5k_omp1_paramslevmar.parameter_flags=Rxy-levmar.parameter_flags=Bfactor-levmar.parameter_flags=Deff-levmar.parameter_flags=Eta-
OMP_NUM_THREADS:=1
OMP_NUM_THREADS:=2
OMP_NUM_THREADS:=4
OMP_NUM_THREADS:=8
OMP_NUM_THREADS:=16
OMP_NUM_THREADS:=32
OMP_NUM_THREADS:=64
Data Set Size:=32k
Skipping 32k
import pandas as pd
import cPickle
str_out=cPickle.load(open('OMP_SOLVER_TIME_5k.pickle', 'rb'))
threads_list = [1,2,4,8,16,32,64]
str_out_sm={}
for k in str_out.keys():
if "strum_5k" in k:
str_out_sm.update({k.replace('levmar.parameter_flags=','').replace('_strum_5k_omp1_params','_'):str_out[k]})
u_dat={}
uniq_ref = set([k.split('_')[1] for k in str_out_sm.keys()])
df_list=[]
for u in uniq_ref:
same_t = lambda: None #Functions are objects, so legit
same_t.strum = []
same_t.eig = []
for t in threads_list:
same_t.strum.append(str_out_sm["omp%d_%s"%(t,u)][0].rsplit(',')[-1].strip()[0:-1])
same_t.eig.append(str_out_sm["omp%d_%s"%(t,u)][1].rsplit(',')[-1].strip()[0:-1])
u_dat.update({u:same_t})
str_E = "EIG_%s"%u
str_S = "STRUM_%s"%u
df_list.append( pd.DataFrame({str_E:same_t.eig,str_S:same_t.strum}, index=threads_list).transpose() )
perf_s = pd.concat(df_list).transpose()
perf_s
# define the figure size and grid layout properties
figsize = (10, 8)
cols = 4
gs = gridspec.GridSpec(cols, cols)
gs.update(hspace=0.6)
fig2 = plt.figure(num=2, figsize=figsize)
fig2.suptitle('5k data', size=16)
ax = []
for i, u in enumerate(uniq_ref):
row = (i // cols)
col = i % cols
ax.append(fig2.add_subplot(gs[row, col]))
if(col==0):
ax[-1].set_ylabel('time [s]')
ax[-1].set_title(str(u)[:-1])
ax[-1].plot(perf_s.index, perf_s['EIG_'+u], 'o', ls='-', ms=6, label='EIG')
ax[-1].plot(perf_s.index, perf_s['STRUM_'+u], 'x', ls='-', ms=6, label='STRUM')
ax[-1].xaxis.set_ticks(threads_list)
ax[-1].set_xscale('log', basex=2)
ax[-1].set_xlim([threads_list[0]/1.8,threads_list[-1]*1.8])
ymin, ymax = ax[-1].get_ylim()
ax[-1].set_ylim([-1,ymax+1])
for label in ax[-1].yaxis.get_ticklabels()[::2]:
label.set_visible(False)
for label in ax[-1].xaxis.get_ticklabels()[1::2]:
label.set_visible(False)
ax[-1].legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.,)
<matplotlib.legend.Legend at 0x7fc4eae1e290>
 |
It may be convenient to submit the run job to a background IPyParallel cluster environment. We can use the default environment, and enable a single python instance to exploit the OMP_NUM_THREADS parallelism of the node.
import ipyparallel as ipp
c = ipp.Client()
rc=c[:]
c.ids #Instance IDs
[0]
By wrapping the solving code from eariler into a function, we can submit this to the background process, and supply the various arguments.
def background_job(list_idx,A_LIST,B_LIST):
str_out={}
import os
threads_list = [64]
for imgs_size in list_idx:
print "Data Set Size:=%s"%imgs_size
#Subselect the smallest data size for now
if imgs_size != "1k":
print "Skipping %s"%imgs_size
continue
for imgs_idx in list_idx[imgs_size]:
A_mat = A_LIST[imgs_idx]; b_vec = B_LIST[imgs_idx]
dat_name = A_mat.split('/')[-1][2:-4]
print "Data Set Name:=%s"%dat_name
#Ensure the A and b data are matched correctly
assert(os.path.dirname(A_mat) == os.path.dirname(b_vec))
for threads in threads_list:
print "OMP_NUM_THREADS:=%d"%threads
val = !OMP_NUM_THREADS={threads} libtbx.python ./OMP_SOLVER.py {A_mat} {b_vec}
key = 'omp' + str(threads) + '_' + dat_name
str_out.update({key:val})
return str_out
Applying the above function to the arguments allows us to grab a handle to the background process, to allow us to determine if and when it has finished. Calling str_out_result.ready() will return True when the funcction has returned, and False otherwise. This result is non-blocking. If however we aim to block until the result is returned, we can use str_out_result.get(), which returns the result for us in any case.
str_out_result=rc.apply_async(background_job,list_idx,A_LIST,B_LIST)
str_out_result.ready()
False
str_out_result.get()
—CLICK FOR OUTPUT—
[{'omp64_strum_1k_omp1_paramslevmar.parameter_flags=Bfactor-': ['individual call time for STRUMPACK: CPU, 298.290s; elapsed, 5.134s',
'individual call time for EIGEN: CPU, 2.620s; elapsed, 0.266s',
'Exiting profiler',
'time for STRUMPACK: CPU, 298.290s; elapsed, 5.134s, #calls: 1',
'time for EIGEN: CPU, 2.620s; elapsed, 0.266s, #calls: 1',
'TOTAL : CPU, 300.910s; elapsed, 5.400s'],
'omp64_strum_1k_omp1_paramslevmar.parameter_flags=Bfactor-levmar.parameter_flags=Deff-': ['individual call time for STRUMPACK: CPU, 400.670s; elapsed, 7.057s',
'individual call time for EIGEN: CPU, 1.000s; elapsed, 0.017s',
'approx_equal eps: 1e-06',
'approx_equal multiplier: 10000000000.0',
'3.5182360272 approx_equal ERROR',
'-3.76297519095 approx_equal ERROR',
'',
'Traceback (most recent call last):',
' File "./OMP_SOLVER.py", line 53, in <module>',
' run_solver(n_rows, n_cols, A_indptr ,A_indices, A_values, b)',
' File "./OMP_SOLVER.py", line 51, in run_solver',
' assert( approx_equal(res_strum.x[i], res_eig.x[i]) )',
'AssertionError',
'Exiting profiler',
'time for STRUMPACK: CPU, 400.670s; elapsed, 7.057s, #calls: 1',
'time for EIGEN: CPU, 1.000s; elapsed, 0.017s, #calls: 1',
'TOTAL : CPU, 401.670s; elapsed, 7.073s'],
'omp64_strum_1k_omp1_paramslevmar.parameter_flags=Bfactor-levmar.parameter_flags=Deff-levmar.parameter_flags=Eta-': ['individual call time for STRUMPACK: CPU, 332.840s; elapsed, 6.318s',
'individual call time for EIGEN: CPU, 2.140s; elapsed, 0.035s',
'approx_equal eps: 1e-06',
'approx_equal multiplier: 10000000000.0',
'3.5182360272 approx_equal ERROR',
'-3.76297519095 approx_equal ERROR',
'',
'Traceback (most recent call last):',
' File "./OMP_SOLVER.py", line 53, in <module>',
' run_solver(n_rows, n_cols, A_indptr ,A_indices, A_values, b)',
' File "./OMP_SOLVER.py", line 51, in run_solver',
' assert( approx_equal(res_strum.x[i], res_eig.x[i]) )',
'AssertionError',
'Exiting profiler',
'time for STRUMPACK: CPU, 332.840s; elapsed, 6.318s, #calls: 1',
'time for EIGEN: CPU, 2.140s; elapsed, 0.035s, #calls: 1',
'TOTAL : CPU, 334.980s; elapsed, 6.353s'],
'omp64_strum_1k_omp1_paramslevmar.parameter_flags=Bfactor-levmar.parameter_flags=Eta-': ['individual call time for STRUMPACK: CPU, 280.900s; elapsed, 5.401s',
'individual call time for EIGEN: CPU, 6.870s; elapsed, 0.275s',
'Exiting profiler',
'time for STRUMPACK: CPU, 280.900s; elapsed, 5.401s, #calls: 1',
'time for EIGEN: CPU, 6.870s; elapsed, 0.275s, #calls: 1',
'TOTAL : CPU, 287.770s; elapsed, 5.676s'],
'omp64_strum_1k_omp1_paramslevmar.parameter_flags=Deff-': ['individual call time for STRUMPACK: CPU, 382.050s; elapsed, 6.886s',
'individual call time for EIGEN: CPU, 0.710s; elapsed, 0.012s',
'approx_equal eps: 1e-06',
'approx_equal multiplier: 10000000000.0',
'-0.527177455789 approx_equal ERROR',
'-3.76297519095 approx_equal ERROR',
'',
'Traceback (most recent call last):',
' File "./OMP_SOLVER.py", line 53, in <module>',
' run_solver(n_rows, n_cols, A_indptr ,A_indices, A_values, b)',
' File "./OMP_SOLVER.py", line 51, in run_solver',
' assert( approx_equal(res_strum.x[i], res_eig.x[i]) )',
'AssertionError',
'Exiting profiler',
'time for STRUMPACK: CPU, 382.050s; elapsed, 6.886s, #calls: 1',
'time for EIGEN: CPU, 0.710s; elapsed, 0.012s, #calls: 1',
'TOTAL : CPU, 382.760s; elapsed, 6.898s'],
'omp64_strum_1k_omp1_paramslevmar.parameter_flags=Deff-levmar.parameter_flags=Eta-': ['individual call time for STRUMPACK: CPU, 340.390s; elapsed, 6.120s',
'individual call time for EIGEN: CPU, 1.560s; elapsed, 0.026s',
'approx_equal eps: 1e-06',
'approx_equal multiplier: 10000000000.0',
'-0.527177455789 approx_equal ERROR',
'-3.76297519095 approx_equal ERROR',
'',
'Traceback (most recent call last):',
' File "./OMP_SOLVER.py", line 53, in <module>',
' run_solver(n_rows, n_cols, A_indptr ,A_indices, A_values, b)',
' File "./OMP_SOLVER.py", line 51, in run_solver',
' assert( approx_equal(res_strum.x[i], res_eig.x[i]) )',
'AssertionError',
'Exiting profiler',
'time for STRUMPACK: CPU, 340.390s; elapsed, 6.120s, #calls: 1',
'time for EIGEN: CPU, 1.560s; elapsed, 0.026s, #calls: 1',
'TOTAL : CPU, 341.950s; elapsed, 6.145s'],
'omp64_strum_1k_omp1_paramslevmar.parameter_flags=Eta-': ['individual call time for STRUMPACK: CPU, 277.570s; elapsed, 5.264s',
'individual call time for EIGEN: CPU, 4.930s; elapsed, 0.081s',
'Exiting profiler',
'time for STRUMPACK: CPU, 277.570s; elapsed, 5.264s, #calls: 1',
'time for EIGEN: CPU, 4.930s; elapsed, 0.081s, #calls: 1',
'TOTAL : CPU, 282.500s; elapsed, 5.344s'],
'omp64_strum_1k_omp1_paramslevmar.parameter_flags=Rxy-': ['individual call time for STRUMPACK: CPU, 438.160s; elapsed, 8.726s',
'individual call time for EIGEN: CPU, 1.060s; elapsed, 0.017s',
'approx_equal eps: 1e-06',
'approx_equal multiplier: 10000000000.0',
'-0.527177455789 approx_equal ERROR',
'-3.76297519095 approx_equal ERROR',
'',
'Traceback (most recent call last):',
' File "./OMP_SOLVER.py", line 53, in <module>',
' run_solver(n_rows, n_cols, A_indptr ,A_indices, A_values, b)',
' File "./OMP_SOLVER.py", line 51, in run_solver',
' assert( approx_equal(res_strum.x[i], res_eig.x[i]) )',
'AssertionError',
'Exiting profiler',
'time for STRUMPACK: CPU, 438.160s; elapsed, 8.726s, #calls: 1',
'time for EIGEN: CPU, 1.060s; elapsed, 0.017s, #calls: 1',
'TOTAL : CPU, 439.220s; elapsed, 8.743s'],
'omp64_strum_1k_omp1_paramslevmar.parameter_flags=Rxy-levmar.parameter_flags=Bfactor-': ['individual call time for STRUMPACK: CPU, 353.900s; elapsed, 6.904s',
'individual call time for EIGEN: CPU, 2.570s; elapsed, 0.042s',
'approx_equal eps: 1e-06',
'approx_equal multiplier: 10000000000.0',
'3.5182360272 approx_equal ERROR',
'-3.76297519095 approx_equal ERROR',
'',
'Traceback (most recent call last):',
' File "./OMP_SOLVER.py", line 53, in <module>',
' run_solver(n_rows, n_cols, A_indptr ,A_indices, A_values, b)',
' File "./OMP_SOLVER.py", line 51, in run_solver',
' assert( approx_equal(res_strum.x[i], res_eig.x[i]) )',
'AssertionError',
'Exiting profiler',
'time for STRUMPACK: CPU, 353.900s; elapsed, 6.904s, #calls: 1',
'time for EIGEN: CPU, 2.570s; elapsed, 0.042s, #calls: 1',
'TOTAL : CPU, 356.470s; elapsed, 6.946s'],
'omp64_strum_1k_omp1_paramslevmar.parameter_flags=Rxy-levmar.parameter_flags=Bfactor-levmar.parameter_flags=Deff-': ['individual call time for STRUMPACK: CPU, 552.200s; elapsed, 11.529s',
'individual call time for EIGEN: CPU, 0.630s; elapsed, 0.030s',
'approx_equal eps: 1e-06',
'approx_equal multiplier: 10000000000.0',
'3.5182360272 approx_equal ERROR',
'-3.76297519095 approx_equal ERROR',
'',
'Traceback (most recent call last):',
' File "./OMP_SOLVER.py", line 53, in <module>',
' run_solver(n_rows, n_cols, A_indptr ,A_indices, A_values, b)',
' File "./OMP_SOLVER.py", line 51, in run_solver',
' assert( approx_equal(res_strum.x[i], res_eig.x[i]) )',
'AssertionError',
'Exiting profiler',
'time for STRUMPACK: CPU, 552.200s; elapsed, 11.529s, #calls: 1',
'time for EIGEN: CPU, 0.630s; elapsed, 0.030s, #calls: 1',
'TOTAL : CPU, 552.830s; elapsed, 11.559s'],
'omp64_strum_1k_omp1_paramslevmar.parameter_flags=Rxy-levmar.parameter_flags=Bfactor-levmar.parameter_flags=Deff-levmar.parameter_flags=Eta-': ['individual call time for STRUMPACK: CPU, 640.470s; elapsed, 13.635s',
'individual call time for EIGEN: CPU, 3.100s; elapsed, 0.051s',
'approx_equal eps: 1e-06',
'approx_equal multiplier: 10000000000.0',
'3.5182360272 approx_equal ERROR',
'-3.76297519095 approx_equal ERROR',
'',
'Traceback (most recent call last):',
' File "./OMP_SOLVER.py", line 53, in <module>',
' run_solver(n_rows, n_cols, A_indptr ,A_indices, A_values, b)',
' File "./OMP_SOLVER.py", line 51, in run_solver',
' assert( approx_equal(res_strum.x[i], res_eig.x[i]) )',
'AssertionError',
'Exiting profiler',
'time for STRUMPACK: CPU, 640.470s; elapsed, 13.635s, #calls: 1',
'time for EIGEN: CPU, 3.100s; elapsed, 0.051s, #calls: 1',
'TOTAL : CPU, 643.570s; elapsed, 13.686s'],
'omp64_strum_1k_omp1_paramslevmar.parameter_flags=Rxy-levmar.parameter_flags=Bfactor-levmar.parameter_flags=Eta-': ['individual call time for STRUMPACK: CPU, 421.790s; elapsed, 7.949s',
'individual call time for EIGEN: CPU, 2.670s; elapsed, 0.044s',
'approx_equal eps: 1e-06',
'approx_equal multiplier: 10000000000.0',
'3.5182360272 approx_equal ERROR',
'-3.76297519095 approx_equal ERROR',
'',
'Traceback (most recent call last):',
' File "./OMP_SOLVER.py", line 53, in <module>',
' run_solver(n_rows, n_cols, A_indptr ,A_indices, A_values, b)',
' File "./OMP_SOLVER.py", line 51, in run_solver',
' assert( approx_equal(res_strum.x[i], res_eig.x[i]) )',
'AssertionError',
'Exiting profiler',
'time for STRUMPACK: CPU, 421.790s; elapsed, 7.949s, #calls: 1',
'time for EIGEN: CPU, 2.670s; elapsed, 0.044s, #calls: 1',
'TOTAL : CPU, 424.460s; elapsed, 7.993s'],
'omp64_strum_1k_omp1_paramslevmar.parameter_flags=Rxy-levmar.parameter_flags=Deff-': ['individual call time for STRUMPACK: CPU, 505.820s; elapsed, 9.863s',
'individual call time for EIGEN: CPU, 1.320s; elapsed, 0.022s',
'approx_equal eps: 1e-06',
'approx_equal multiplier: 10000000000.0',
'-0.527177455789 approx_equal ERROR',
'-3.76297519095 approx_equal ERROR',
'',
'Traceback (most recent call last):',
' File "./OMP_SOLVER.py", line 53, in <module>',
' run_solver(n_rows, n_cols, A_indptr ,A_indices, A_values, b)',
' File "./OMP_SOLVER.py", line 51, in run_solver',
' assert( approx_equal(res_strum.x[i], res_eig.x[i]) )',
'AssertionError',
'Exiting profiler',
'time for STRUMPACK: CPU, 505.820s; elapsed, 9.863s, #calls: 1',
'time for EIGEN: CPU, 1.320s; elapsed, 0.022s, #calls: 1',
'TOTAL : CPU, 507.140s; elapsed, 9.884s'],
'omp64_strum_1k_omp1_paramslevmar.parameter_flags=Rxy-levmar.parameter_flags=Deff-levmar.parameter_flags=Eta-': ['individual call time for STRUMPACK: CPU, 400.650s; elapsed, 7.382s',
'individual call time for EIGEN: CPU, 2.570s; elapsed, 0.042s',
'approx_equal eps: 1e-06',
'approx_equal multiplier: 10000000000.0',
'-0.527177455789 approx_equal ERROR',
'-3.76297519095 approx_equal ERROR',
'',
'Traceback (most recent call last):',
' File "./OMP_SOLVER.py", line 53, in <module>',
' run_solver(n_rows, n_cols, A_indptr ,A_indices, A_values, b)',
' File "./OMP_SOLVER.py", line 51, in run_solver',
' assert( approx_equal(res_strum.x[i], res_eig.x[i]) )',
'AssertionError',
'Exiting profiler',
'time for STRUMPACK: CPU, 400.650s; elapsed, 7.382s, #calls: 1',
'time for EIGEN: CPU, 2.570s; elapsed, 0.042s, #calls: 1',
'TOTAL : CPU, 403.220s; elapsed, 7.424s'],
'omp64_strum_1k_omp1_paramslevmar.parameter_flags=Rxy-levmar.parameter_flags=Eta-': ['individual call time for STRUMPACK: CPU, 440.060s; elapsed, 8.054s',
'individual call time for EIGEN: CPU, 2.350s; elapsed, 0.038s',
'approx_equal eps: 1e-06',
'approx_equal multiplier: 10000000000.0',
'-0.527177455789 approx_equal ERROR',
'-3.76297519095 approx_equal ERROR',
'',
'Traceback (most recent call last):',
' File "./OMP_SOLVER.py", line 53, in <module>',
' run_solver(n_rows, n_cols, A_indptr ,A_indices, A_values, b)',
' File "./OMP_SOLVER.py", line 51, in run_solver',
' assert( approx_equal(res_strum.x[i], res_eig.x[i]) )',
'AssertionError',
'Exiting profiler',
'time for STRUMPACK: CPU, 440.060s; elapsed, 8.054s, #calls: 1',
'time for EIGEN: CPU, 2.350s; elapsed, 0.038s, #calls: 1',
'TOTAL : CPU, 442.410s; elapsed, 8.093s']}]
str_out=str_out_result.get()
import json, cPickle
with open("OMP_SOLVER_TIME_1k_OMP64_ASYNC.json", "w") as OMP_SOLVER_TIME_FILE:
OMP_SOLVER_TIME_FILE.write(json.dumps(str_out))
with open("OMP_SOLVER_TIME_1k_OMP64_ASYNC.pickle", "w") as OMP_SOLVER_TIME_FILE:
OMP_SOLVER_TIME_FILE.write(cPickle.dumps(str_out))
We can now submit a large background job, and close the window until it has completed. This will run asynchronously on the cluster backend, and allow us to poll it upon our return.
def background_job_10k(list_idx,A_LIST,B_LIST):
str_out={}
import os
threads_list = [1,2,4,8,16,32]
for imgs_size in list_idx:
print "Data Set Size:=%s"%imgs_size
#Subselect the smallest data size for now
if imgs_size != "10k":
print "Skipping %s"%imgs_size
continue
for imgs_idx in list_idx[imgs_size]:
A_mat = A_LIST[imgs_idx]; b_vec = B_LIST[imgs_idx]
dat_name = A_mat.split('/')[-1][2:-4]
print "Data Set Name:=%s"%dat_name
#Ensure the A and b data are matched correctly
assert(os.path.dirname(A_mat) == os.path.dirname(b_vec))
for threads in threads_list:
print "OMP_NUM_THREADS:=%d"%threads
val = !OMP_NUM_THREADS={threads} libtbx.python ./OMP_SOLVER.py {A_mat} {b_vec}
key = 'omp' + str(threads) + '_' + dat_name
str_out.update({key:val})
return str_out
str_out_result=rc.apply_async(background_job_10k,list_idx,A_LIST,B_LIST)
str_out_result.ready()
False